Development News Brief
Get Galaxy
new: % hg clone http://www.bx.psu.edu/hg/galaxy galaxy-dist
upgrade: % hg pull -u -r ec29ce8e27a1
# GCC2012
The 2012 Galaxy Community Conference (GCC2012) is a few short days away. Late registration is still open.
# FreeBayes Migration
The tool Human Genome Variation -> Freebayes has moved from the Galaxy distribution to the Galaxy Main Tool Shed.
FreeBayes
is a high-performance, flexible, and open-source Bayesian genetic variant detector. It operates on BAM alignment files, which are produced by most contemporary short-read aligners.
# EMBOSS Update
The emboss_5 repository in the Galaxy Main Tool Shed has been updated to include information for automatically installing the EMBOSS Version 5.0.0 tool dependencies. If you've installed the emboss_5 repository into your local Galaxy instance, get this update from the tool shed, and you'll have the option of installing the tool dependencies.
# Admin Genome Indexing
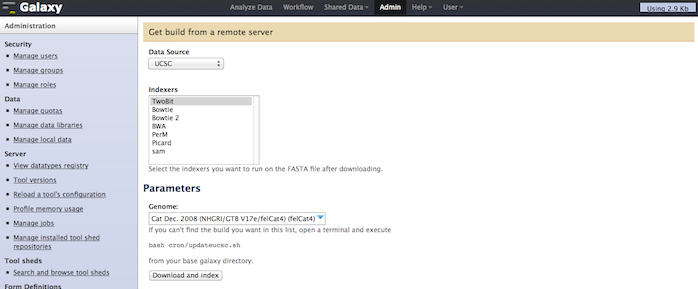
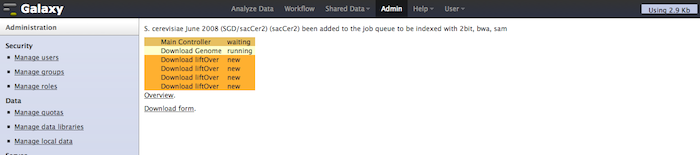
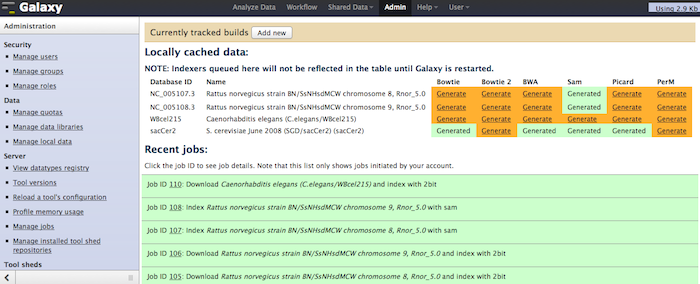
If enable_beta_job_managers = True, Galaxy will now show a new option in the admin interface, titled Manage Local Data. With this tool, a Galaxy admin will be able to select and download any genome already contained with the local $GALAXYROOT/tool-data/shared/ucsc/builds.txt file sourced from UCSC, NCBI, or Ensembl, optionally indexing the FASTA data with one or more of BWA, Bowtie, Bowtie 2, PerM, Picard, and SAM. After these processes finish, the indexed build will then be available for other tools in the analysis section. This does not replace setting up a build.txt file (read more ...). This would replace many of the manual indexing processes for commonly used tools ([read more...](/admin/NGS Local Setup/)). Please this tool is still beta, feedback and bug reports welcome at galaxy-dev@bx.psu.edu for discussion. We will also touch on it during a breakout session at GCC (Day 2, Section 8, Automation Strategies).
Example of selecting a source, the indexes to create, and the target genome:
The status of the indexing for a single genome:
And a global status of all indexes for all genomes in progress:
# Simplified install of 3rd-party Dependencies
Automatic third-party tool dependency installation and compilation with installed repositories
Tool shed repository owners can define information in their repositories that enable third party tool dependencies to be automatically installed along with the repository for those repositories that contain tools that require the dependencies. The process for enabling this is the inclusion of a simple xml file named tool_dependencies.xml in the repository. Read more…
# Improved Error Handling
Several changes made in in determining errors from tool exit codes and output. In the past, Galaxy has determined if a tool has an error by checking if anything was written to standard error (stderr}). Now each tool's XML can be customized to check the tool's exit code, standard output (stdout), and standard error (stderr). The exit code can be checked against a supplied range of values, and regular expressions can be checked against stdout and stderr. Users can specify if the error should result in a warning or a fatal error. If this feature is not specified, then the previous behavior of just checking stderr will be used.
# Tools
Admin/Config/Tool Dependencies
-
Tophat2 wrapper enhancements:
- use Bowtie2 build
- add option to report discordant pairs
- update tests
- add Bowtie2 preset options
# User Interface (UI)
-
Enhancements
-
Full integration with myExperiment website. Galaxy workflows can be
- exported from Galaxy to myExperiment and
- searched and imported from myExperiment website
- Support for genomes as first-class objects
-
# Galaxy Track Browser (GTB)
- New parameter space visualization for Trackster
- Make bookmarks available in shared Trackster visualizations
# Tool Shed
-
Enhancements
- The tool shed's category grid is now displayed when searching and browsing tool sheds from a local Galaxy instance.
- This category grid's search feature searches valid repository names and descriptions when browsing a tool shed from Galaxy.
- Going forward from this Galaxy release, the main Galaxy tool shed will track the Galaxy releases and the galaxy-dist repository on bitbucket.
# Bug Fixes * Support Unvalidated values when exporting histories
# Announcements
Collaboration
The new RGalaxy package in Bioconductor. Read more …
GalaxyCzars
The [GalaxyCzars group](/community/galaxy-admins/) was launched and had its [first meetup on July 9](/community/galaxy-admins/Meetups/2012-07-09/). GalaxyCzars is a group of people that manage large local Galaxy installations. See the [meeting writeup](/community/galaxy-admins/Meetups/2012-07-09/) for links to slides and a screencast. The GalaxyCzars will have a breakout at [GCC2012](/events/gcc2012/), and the next conference call will follow in September.## Galaxy is Hiring!
Want to work on one of the fastest growing open source bioinformatics projects around? The [Galaxy Project](http://galaxyproject.org/), a highly successful high throughput data analysis platform for Life Sciences with over 15,000 users worldwide, is hiring. [Read more...](/galaxy-is-hiring/)
----
# About Galaxy
The GalaxyTeam is a part of BX at Penn State, and the Biology and Mathematics and Computer Science departments at Emory University.
Galaxy is supported in part by NSF, NHGRI, the Huck Institutes of the Life Sciences, and The Institute for CyberScience at Penn State, and Emory University.
Join us at Twitter @galaxyproject or just read our tweets Galaxy on Twitter



